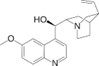
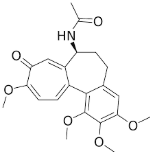
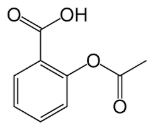
Structural Analogs and Derivatives of Natural Products in Modern Therapeutics
August 05, 2025
This paper presents an exploration of natural product derivatives in modern therapeutics
I. Introduction: From Nature to Laboratory
The journey from natural compounds à therapeutic innovations represent pharmacology's profound achievements.
Natural products have provided critical scaffolds for drug development across the spectrum of human disease, with ~ 60% of approved pharmaceuticals deriving from or being inspired by natural sources.
Element 1: Molecular Evolution Timeline
A molecular tree displaying the chronological development of natural product-derived therapeutics.
Navigate from primitive antimicrobials like penicillin (1928) through plant alkaloids to modern biologics & gene therapies.
Zoom-in to reveal molecular structures with increasing detail of therapeutic classes (antibiotics, chemotherapeutics, etc.)
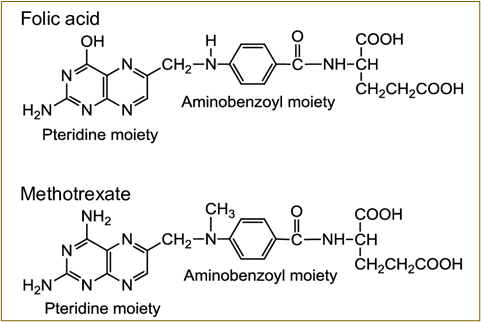
II. Chemically Synthesized Small-Molecule Drugs: Structural Modifications and Therapeutic Implications
The evolution from folic acid to methotrexate exemplifies rational drug design based on natural templates.
Element 2: Comparative Structure
Nucleoside Analogs: Mechanism of Action Visualization
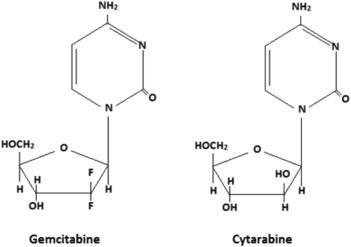
Cytarabine (Ara-C) and gemcitabine incorporate into DNA as false pyrimidine bases, leading to chain termination during replication—a mechanism uniquely effective against rapidly dividing cancer cells.
Element 3: DNA Replication Disruption showing DNA polymerase interaction with nucleoside analogs
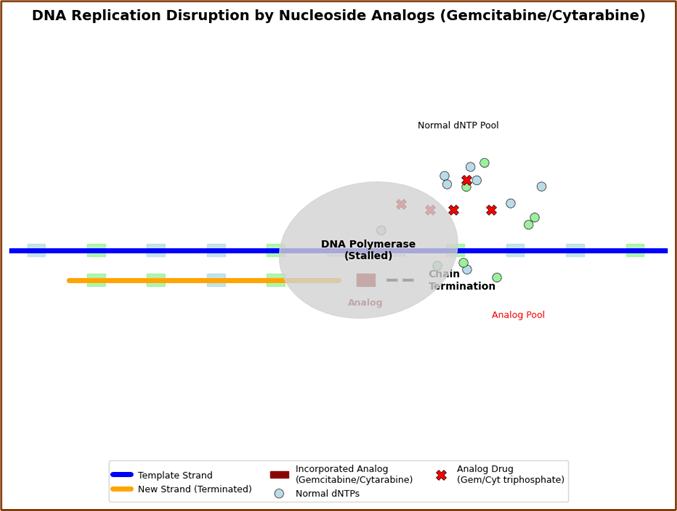
Table: Illustrations of few Natural Product-Derived Therapeutics
Drug | Structure | Natural Origin | Structural Modification | Clinical Pharmacokinetics | Therapeutic Applications | Molecular Targets |
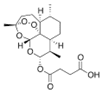
Paclitaxel |
| Taxus brevifolia (Pacific yew) bark | Semi-synthetic esterification; C-13 side chain modification | t½: 5.8h; 89-98% protein bound; hepatic metabolism | Ovarian, breast, lung carcinomas; Kaposi's sarcoma | β-tubulin stabilization |
Vincristine |
| Catharanthus roseus (Madagascar periwinkle) | Alkaloid isolation; C-3 esterification | t½: 85h; 75% protein bound; biliary excretion | ALL, Hodgkin's lymphoma, neuroblastoma | Tubulin polymerization inhibition |
Doxorubicin |
| Streptomyces peucetius | Anthracycline glycosylation; C-14 hydroxylation | t½: 20-48h; 74- 76% protein bound; hepatic metabolism | Solid tumors, leukemias, lymphomas | Topoisomerase II inhibition; DNA intercalation |
Etoposide |
| Podophyllum peltatum (American mandrake) | Semi-synthetic glycosylation; 4'- demethylepipodophyllotoxin derivative | t½: 6h; 97% protein bound; hepatic metabolism | SCLC, testicular cancer, lymphomas | Topoisomerase II inhibition |
Artesunate |
| Artemisia annua (Sweet wormwood) leaves | Semi-synthetic ester derivative of Artemisinin | Prodrug; Rapidly hydrolyzed to active Dihydroartemisinin (DHA, t½ ~1h); metabolized (glucuronidation) | Severe malaria, uncomplicated malaria (combination therapy) | Heme activation -> free radicals; alkylation of parasite proteins (e.g., PfATP6) |
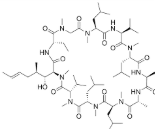
Cyclosporine (Ciclosporin) |
| Tolypocladium inflatum (fungus) | Isolated natural product | t½: ~19h (variable); >90% protein bound; hepatic metabolism (CYP3A4), biliary excretion | Organ transplant rejection prevention; Rheumatoid arthritis, Psoriasis | Calcineurin inhibition (via cyclophilin binding), reduces IL-2 gene transcription |
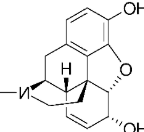
Morphine |
| Papaver somniferum (Opium poppy) latex | Isolated alkaloid; used as salt (e.g., sulfate) | t½: 2-3h; 30-40% protein bound; hepatic metabolism (glucuronidation to M6G/M3G), renal excretion | Severe acute and chronic pain | Mu (μ)-opioid receptor agonist (primary); also, kappa/delta |
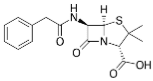
Penicillin G (Benzylpenicillin) | | Penicillium species (fungus) | Natural product from fermentation; used as salt (K+, Na+) | t½: ~30 min; ~60% protein bound; rapid renal excretion (tubular secretion) | Susceptible bacterial infections (Strep., Syphilis, Meningitis) | Penicillin-Binding Proteins (PBPs) inhibition, blocks cell wall synthesis |
Lovastatin |
| Aspergillus terreus (fungus) | Isolated natural product (prodrug) | Prodrug hydrolyzed to active acid (t½ ~2-3h); >95% protein bound; extensive hepatic first-pass metabolism (CYP3A4) | Hypercholesterolemia, cardiovascular risk reduction | HMG-CoA reductase inhibition |
Irinotecan |
| Camptotheca acuminata (Happy Tree) bark/stem | Semi-synthetic derivative of Camptothecin (prodrug) | Prodrug (t½ ~6-12h) converted to active SN-38 (t½ ~10-20h); high protein binding (SN-38 >95%); hepatic metabolism (CES, UGT1A1), biliary excretion | Metastatic colorectal cancer | Topoisomerase I inhibition (SN-38 stabilizes cleavable complex) |
Erythromycin |
| Saccharopolyspora erythraea (bacterium) | Isolated natural product | t½: ~1.5-2h; 70-90% protein bound; hepatic metabolism (CYP3A4 demethylation), primarily biliary excretion | Respiratory tract infections, skin infections, chlamydia | Bacterial 50S ribosomal subunit binding, inhibits protein synthesis |
Galantamine |
| Galanthus species (Snowdrop bulbs) | Isolated alkaloid or synthetic | t½: ~7h; ~18% protein bound; hepatic metabolism (CYP2D6/3A4), renal excretion | Mild to moderate Alzheimer's disease | Acetylcholinesterase (AChE) inhibition; Allosteric modulation of nicotinic receptors |
Quinine |
| Cinchona spp. bark | Alkaloid isolation | t½: 11-18h; 70-95% protein bound; hepatic (CYP3A4) | Malaria (chloroquine-resistant), nocturnal leg cramps | Hemozoin biocrystallization inhibition in Plasmodium |
Digoxin |
| Digitalis lanata (foxglove) | Isolated cardiac glycoside | t½: 36-48h; 20-30% protein bound; renal excretion | Heart failure, atrial fibrillation | Na+/K+ ATPase inhibition → increased intracellular Ca²⁺ |
Colchicine |
| Colchicum autumnale (autumn crocus) | Alkaloid isolation | t½: 26-31h; 30-50% protein bound; hepatic metabolism | Gout, familial Mediterranean fever | Tubulin depolymerization → inhibition of inflammasome NLRP3 |
Aspirin |
| Salix alba (willow bark) | Acetylation of salicylic acid | t½: 15-20min (aspirin) → salicylic acid (t½: 2-3h); hepatic | Pain, inflammation, antiplatelet therapy | COX-1/COX-2 inhibition → reduced prostaglandin synthesis |
III. Biologics and Recombinant Proteins: Engineering Natural Templates
Element 4: Insulin Structure profiles
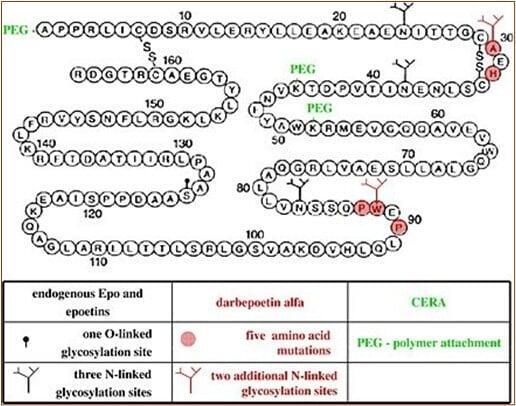
Erythropoietin (EPO): Glycoengineering for Enhanced Pharmacokinetics
The strategic amplification of N-linked glycosylation sites in darbepoetin alfa from 3 to 5 demonstrates advanced glycoengineering to reduce renal clearance and extend serum half-life.
Element 5: Glycosylation Pattern Comparison
IV. Monoclonal Antibodies (mAbs): Structure-Function Relationships
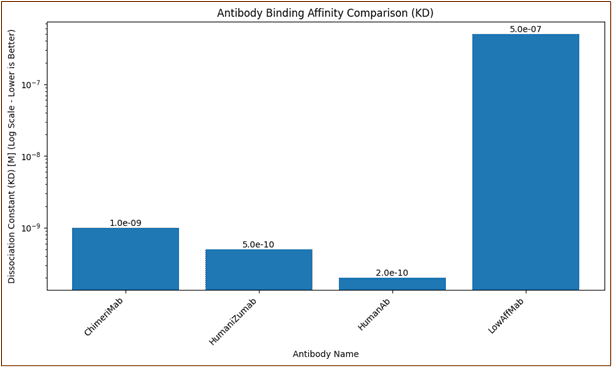
Tumor-Targeting mAbs: Engineering Specificity and Functionality
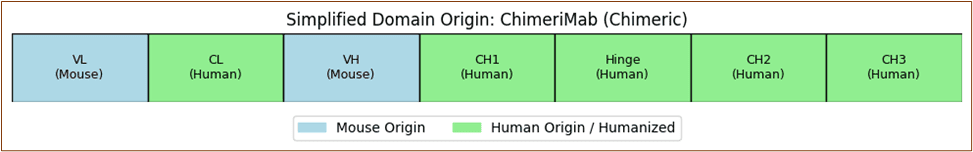
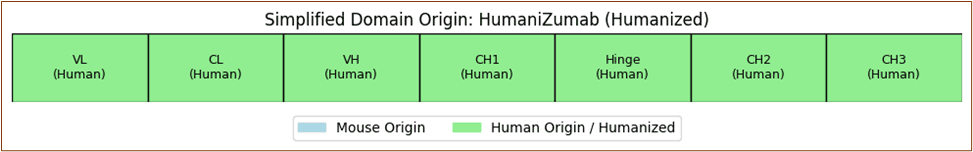
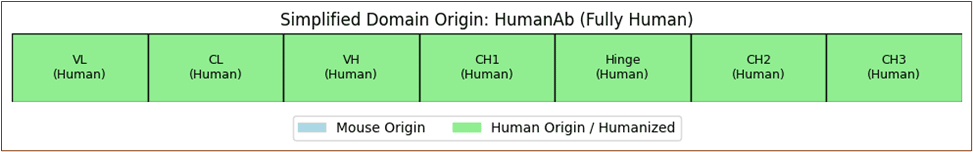
Rituximab's chimeric structure merges murine variable regions with human constant domains to optimize target specificity while minimizing immunogenicity.
The resulting antibody leverages natural complement activation and Fc receptor recognition pathways.
Element 6: Antibody Structure-Function Exploration
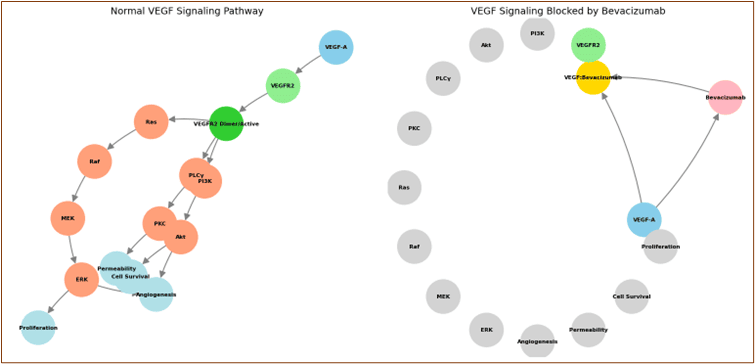
Bevacizumab: Angiogenesis Inhibition Mechanism
The structural mimicry of the endogenous VEGF receptor binding domain in bevacizumab demonstrates biomimetic drug design principles at the protein level.
Element 7: VEGF Signaling Blockade
V. Vaccines: Biomimetic Delivery Systems
Lipid Nanoparticle-Encapsulated mRNA Vaccines
The innovative encapsulation of nucleoside-modified mRNA in ionizable lipid nanoparticles mimics viral envelope fusion mechanisms while avoiding integration risks.
Element 8: mRNA Vaccine Delivery Simulation
Implementation: Step-by-step animation of lipid nanoparticle entry, endosomal escape, and mRNA translation.
Key features:
Interactive click-through progression of delivery stages.
Magnified view of lipid phase transitions during endosomal acidification.
Real-time visualization of ribosomal translation and protein synthesis.
Immune response pathway activation showing antigen presentation and antibody production.
Adenoviral Vectors: Natural Viral Machinery Repurposed
The ChAdOx1 nCoV-19 vaccine capitalizes on natural adenovirus cellular tropism while engineering removal of replication capability.
Element 9: Adenoviral Vector Engineering Visualization
Implementation: Cross-sectional model of modified adenovirus structure with elements showing genetic modifications.
Key features: Toggle between wild-type and vaccine vector genome composition.
Animated cell entry and nuclear transport simulation.
DNA transcription visualization without replication capability.
Dendritic cell presentation pathway with T-cell activation simulation.
VI. Cell and Gene Therapies: Reprogramming Natural Systems
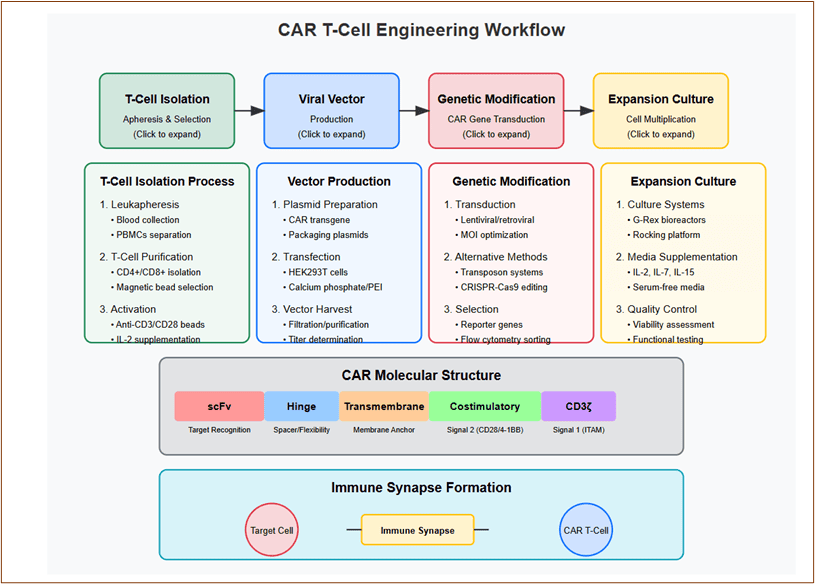
CAR T-Cells: Synthetic Biology Meets Cellular Immunity
The fusion of natural T-cell receptor signaling domains with synthetic tumor-targeting scFv regions in Chimeric Antigen Receptors (CARs) represents the intersection of protein engineering and cellular immunotherapy.
Element 10: CAR T-Cell Engineering and Function
Workflow showing T-cell isolation through engineering to tumor targeting.
Key features: Process steps revealing protocols and cellular modifications.
CAR structural components and assembly.
Immune synapse formation between CAR T-cell and tumor cell.
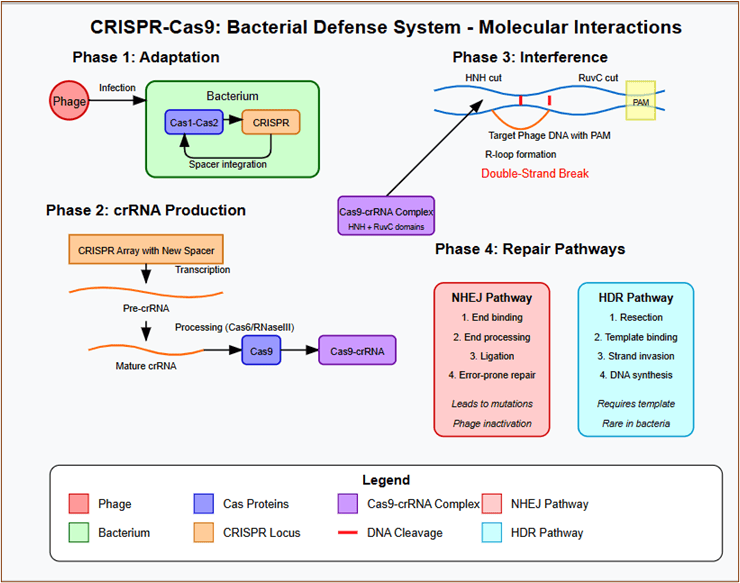
CRISPR-Cas9: Bacterial Defence Systems Repurposed
The adaptation of Streptococcus pyogenes CRISPR systems for therapeutic genome editing exemplifies the translation of natural bacterial defence mechanisms into precision medicine tools.
Element 11: CRISPR-Cas9 Mechanism and Engineering
Key features:
DNA binding, R-loop formation, and cleavage.
Engineered variants with expanded PAM recognition.
Side-by-side comparison of NHEJ and HDR repair pathway outcomes.
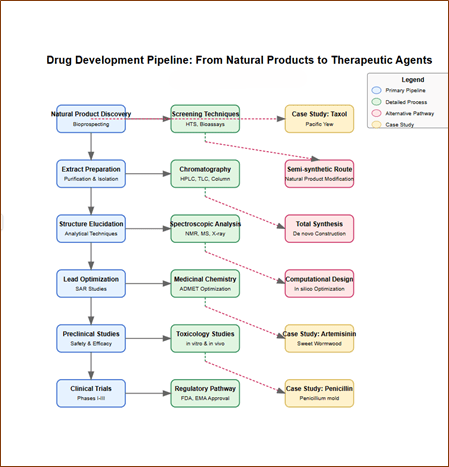
VII. Drug Development Pipeline: From Natural Products to Therapeutic Agents
Element 12: Comprehensive Development Workflow
Implementation: Multi-branched interactive flowchart with expandable process nodes and case studies.
Key features: Primary pathway from natural product identification through clinical approval
Sub-processes revealing methodologies.
Alternative pathway branches showing semi-synthetic and fully synthetic approaches.
Case study examples.
VIII. Future Directions and Challenges
The ongoing evolution of natural product-inspired therapeutics faces challenges in bioavailability, synthesis complexity, and scalability.
Emerging technologies in AI-driven discovery, synthetic biology, and computational design promise to accelerate innovation in this field.
Element 13: Future Technologies Exploration
https://claude.ai/public/artifacts/d3047fb7-673b-436c-b58a-0b29e8718b7d
Digital implementation: Interactive forecast visualization of emerging methodologies and their potential impact.
Key features: Timeline projection of key technologies through 2030.
Impact assessment matrices for various therapeutic categories.
Expert commentary video clips accessible through embedded links.
Interactive surveys capturing user predictions and expertise.
IX. Accessibility and User Experience Considerations
Color schemes for vision accessibility.
Multiple information modalities (visual, textual, numerical) for diverse learning preferences.
Downloadable versions for educational use.
Mobile-responsive design for cross-platform compatibility.
X. Conclusion: Nature as Template and Teacher
The evolution of natural product-derived therapeutics demonstrates the profound utility of biological templates in drug design.
From simple structural analogs to complex cellular therapies, the pharmaceutical landscape continues to draw inspiration from nature's molecular architecture.
As computational tools, synthetic methods, and biological understanding advance, the boundary between natural and engineered therapeutics increasingly blurs, promising ever more sophisticated and effective treatment modalities.
Element 14: Evolutionary Tree of Therapeutic Innovation
Implementation: Interactive visualization showing the branching development of therapeutic modalities from common natural origins.
Key features: Chronological progression from early natural product isolation to advanced engineering.
Convergent and divergent evolutionary pathways in therapeutic development.
Predictive branches suggesting future development directions Integration of all previously explored therapeutic categories into a unified conceptual framework
XI. Points to ponder
Prominent observations that add value to the above discussed content are considered as below:
Arrangement of funding or investment plan for natural product-based innovation as complexity is high.
Regulatory hurdles for modified natural products.
Pathway for clinical trials that can be performed effectively to obtain expedited entry in the market.
Promotion of open access to natural product libraries for guidance.
XII. Acknowledgement & Disclosure statement
XIII. Major Reference Links
10.1186/s40064-015-1493-6
We, at Isazi; encourage, appreciate & support action on the fundamental scientific aspects that lay the foundation of applied sciences.